Multiple ensemble diagnostic regression (MDER) for constraining future austral jet position
Overview
Wenzel et al. (2016) use multiple ensemble diagnostic regression (MDER) to constrain the CMIP5 future projection of the summer austral jet position with several historical process-oriented diagnostics and respective observations.
The following plots are reproduced:
Absolute correlation between the target variable and the diagnostics.
Scatterplot between the target variable and the MDER-calculated linear combination of diagnostics.
Boxplot of RMSE for the unweighted multi-model mean and the (MDER) weighted multi-model mean of the target variable in a pseudo-reality setup.
Time series of the target variable for all models, observations and MDER predictions.
Errorbar plots for all diagnostics.
Scatterplots between the target variable and all diagnostics.
Available recipes and diagnostics
Recipes are stored in recipes/
recipe_wenzel16jclim.yml
Diagnostics are stored in diag_scripts/
austral_jet/asr.ncl
austral_jet/main.ncl
mder/absolute_correlation.ncl
mder/regression_stepwise.ncl
mder/select_for_mder.ncl
User settings in recipe
Preprocessor
extract_region: Region extraction.extract_levels: Pressure level extraction.area_statistics: Spatial average calculations.
Script austral_jet/asr.ncl
season, str: Season.average_ens, bool, optional (default:False): Average over all given ensemble members of a climate model.wdiag, array of str, optional: Names of the diagnostic for MDER output. Necessary when MDER output is desired.wdiag_title, array of str, optional: Names of the diagnostic in plots.
Script austral_jet/main.ncl
styleset, str: Style set used for plotting the multi-model plots.season, str: Season.average_ens, bool, optional (default:False): Average over all given ensemble members of a climate model.rsondes, array of str, optional: Additional observations used in the plot but not for MDER output.rsondes_file, array of str, optional: Paths to the additional observations Necessary whenrsondesis given.rsondes_yr_min, int, optional: Minimum year for additional observations. Necessary whenrsondesis given.rsondes_yr_max, int, optional: Maximum year for additional observations. Necessary whenrsondesis given.wdiag, array of str, optional: Names of the diagnostic for MDER output. Necessary when MDER output is desired.wdiag_title, array of str, optional: Names of the diagnostic in plots.derive_var, str, optional: Derive variables using NCL functions. Must be one of"tpp","mmstf".derive_latrange, array of float, optional: Latitude range for variable derivation. Necessary ifderive_varis given.derive_lev, float, optional: Pressure level (given in Pa) for variable derivation. Necessary ifderive_varis given.
Script mder/absolute_correlation.ncl
p_time, array of int: Start years for future projections.p_step, int: Time range for future projections (in years).scal_time, array of int: Time range for base period (in years) for anomaly calculations used whencalc_type = "trend".time_oper, str: Operation used in NCLtime_operationfunction.time_opt, str: Option used in NCLtime_operationfunction.calc_type, str: Calculation type for the target variable. Must be one of"trend","pos","int".domain, str: Domain tag for provenance tracking.average_ens, bool, optional (default:False): Average over all given ensemble members of a climate model.region, str, optional: Region used for area aggregation. Necessary if input of target variable is multidimensional.area_oper, str, optional: Operation used in NCLarea_operationfunction. Necessary if multidimensional is given.plot_units, str, optional (attribute forvariable_info): Units for the target variable used in the plots.
Script mder/regression_stepwise.ncl
p_time, array of int: Start years for future projections.p_step, int: Time range for future projections (in years).scal_time, array of int: Time range for base period (in years) for anomaly calculations used whencalc_type = "trend".time_oper, str: Operation used in NCLtime_operationfunction.time_opt, str: Option used in NCLtime_operationfunction.calc_type, str: Calculation type for the target variable. Must be one of"trend","pos","int".domain, str: Domain tag for provenance tracking.average_ens, bool, optional (default:False): Average over all given ensemble members of a climate model.smooth, bool, optional (default:False): Smooth time period with 1-2-1 filter.iter, int, optional: Number of iterations for smoothing. Necessary whensmoothis given.cross_validation_mode, bool, optional (default:False): Perform cross-validation.region, str, optional: Region used for area aggregation. Necessary if input of target variable is multidimensional.area_oper, str, optional: Operation used in NCLarea_operationfunction. Necessary if multidimensional is given.plot_units, str, optional (attribute forvariable_info): Units for the target variable used in the plots.
Script mder/select_for_mder.ncl
wdiag, array of str: Names of the diagnostic for MDER output. Necessary when MDER output is desired.domain, str: Domain tag for provenance tracking.ref_dataset, str: Style set used for plotting the multi-model plots.average_ens, bool, optional (default:False): Average over all given ensemble members of a climate model.derive_var, str, optional: Derive variables using NCL functions. Must be one of"tpp","mmstf".
Variables
ta (atmos, monthly, longitude, latitude, pressure level, time)
uajet (atmos, monthly, time)
va (atmos, monthly, longitude, latitude, pressure level, time)
ps (atmos, monthly, longitude, latitude, time)
asr (atmos, monthly, longitude, latitude, time)
Observations and reformat scripts
ERA-Intermin (ta, uajet, va, ps)
CERES-EBAF (asr)
References
Wenzel, S., V. Eyring, E.P. Gerber, and A.Y. Karpechko: Constraining Future Summer Austral Jet Stream Positions in the CMIP5 Ensemble by Process-Oriented Multiple Diagnostic Regression. J. Climate, 29, 673–687, doi:10.1175/JCLI-D-15-0412.1, 2016.
Example plots

Fig. 107 Time series of the the target variable (future austral jet position in the RCP 4.5 scenario) for the CMIP5 ensemble, observations, unweighted multi-model mean projections and (MDER) weighted multi-model mean projections.

Fig. 108 Scatterplot of the target variable (future austral jet position in the RCP 4.5 scenario) vs. the MDER-determined linear combination of diagnostics for the CMIP5 ensemble.

Fig. 109 Boxplot for the RMSE of the target variable for the unweighted and (MDER) weighted multi-model mean projections in a pseudo-reality setup.
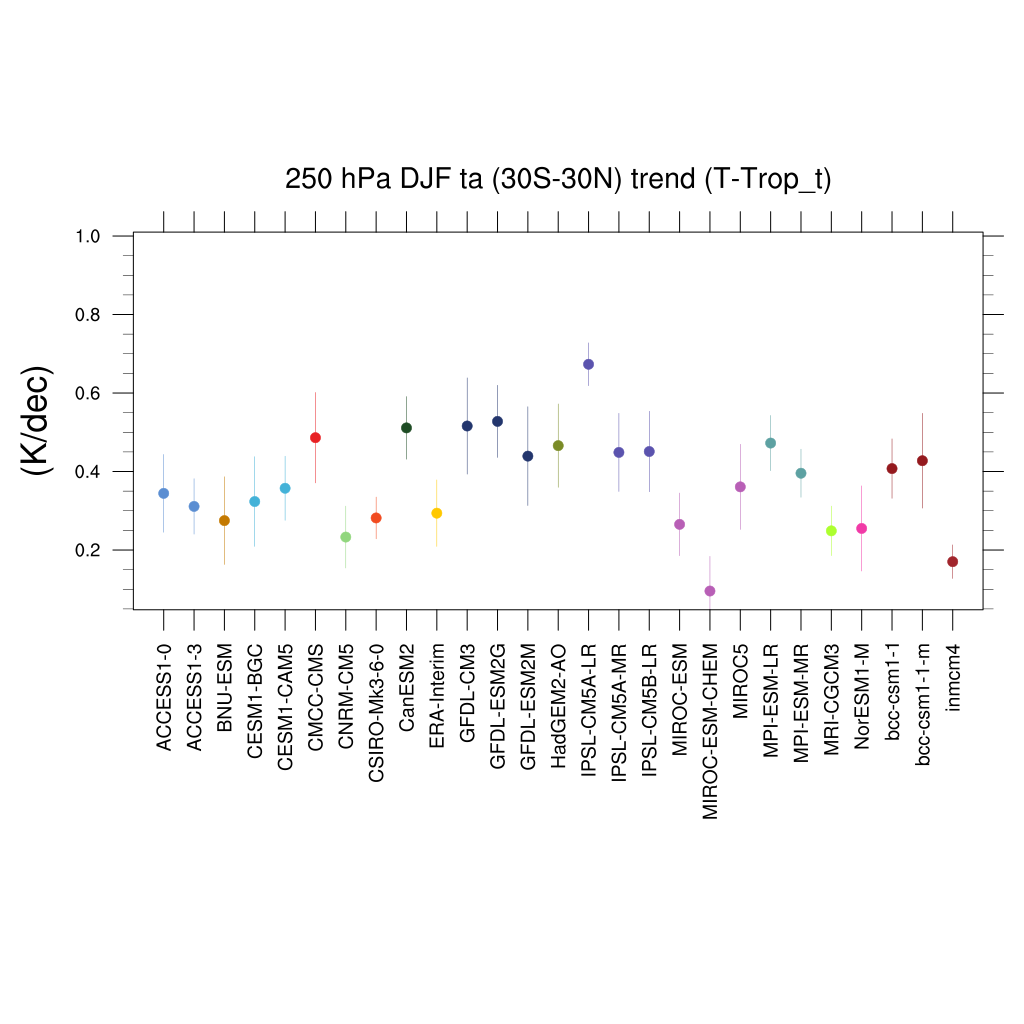
Fig. 110 Trends in tropical DJF temperature at 250hPa for different CMIP5 models and observations.
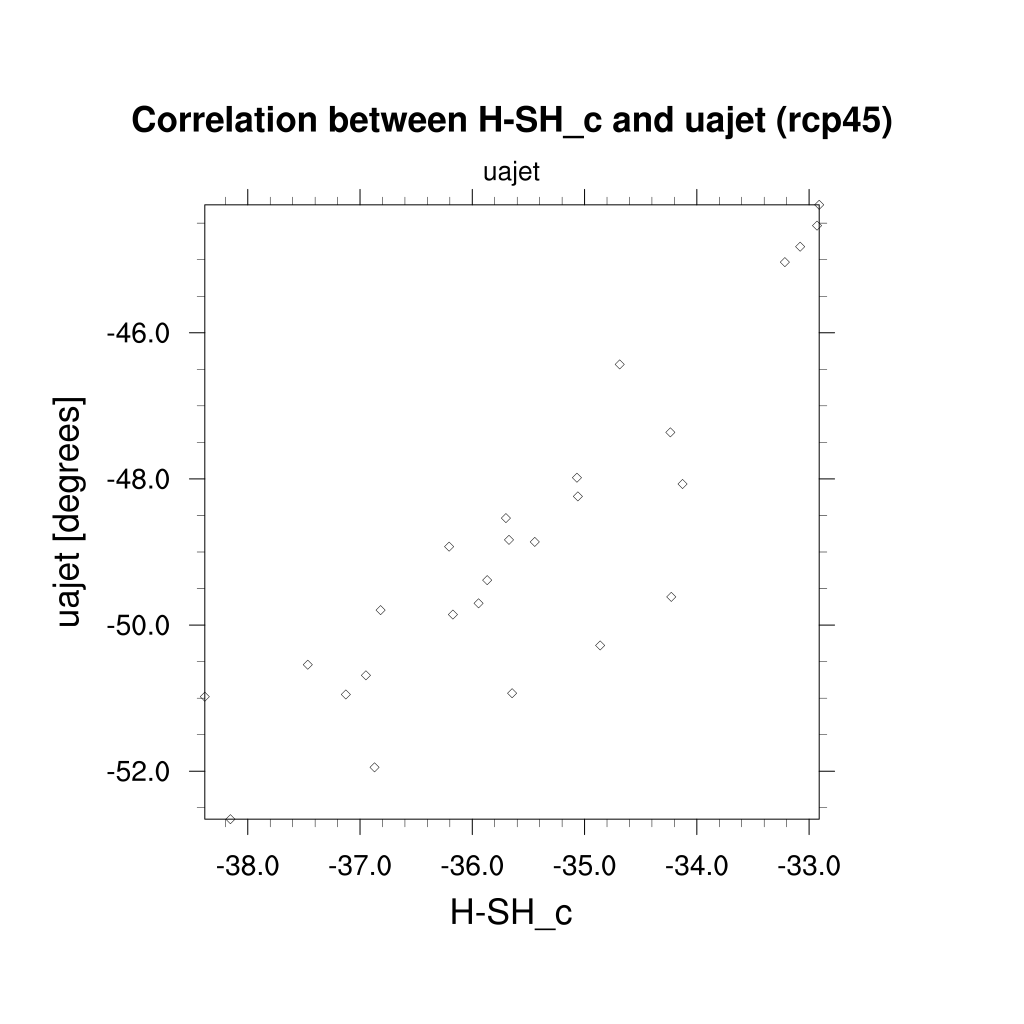
Fig. 111 Scatterplot of the target variable (future austral jet position in the RCP 4.5 scenario) vs. a single diagnostic, the historical location of the Southern hemisphere Hadley cell boundary for the CMIP5 ensemble.