Loading, processing, and visualizing data#
This notebook shows how to load a dataset, use the preprocessor functions, and visualize the result. In this notebook we will plot the annual mean temperature from 1850 till 2100 from one model.
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
import iris.quickplot
from esmvalcore.config import CFG
from esmvalcore.dataset import Dataset
from esmvalcore.esgf import download, ESGFFile
from esmvalcore.preprocessor import area_statistics, annual_statistics
Configure ESMValCore so it searches the ESGF for data
[2]:
CFG['search_esgf'] = 'when_missing'
Define the dataset we are going to use. In this case surface air temperature (tas).
[3]:
tas = Dataset(
short_name='tas',
mip='Amon',
project='CMIP5',
dataset='MPI-ESM-MR',
ensemble='r1i1p1',
exp='historical',
timerange='1850/2000',
)
tas
[3]:
Dataset:
{'dataset': 'MPI-ESM-MR',
'project': 'CMIP5',
'mip': 'Amon',
'short_name': 'tas',
'ensemble': 'r1i1p1',
'exp': 'historical',
'timerange': '1850/2000'}
In order to compute the area average later on, we add the cell areas (areacella) as a supplementary dataset. This will append a new dataset to the list of supplementary datasets. Its facets are copied from the tas dataset and updated with the provided facets:
[4]:
tas.add_supplementary(short_name='areacella', mip='fx', ensemble='r0i0p0')
tas.supplementaries
[4]:
[Dataset:
{'dataset': 'MPI-ESM-MR',
'project': 'CMIP5',
'mip': 'fx',
'short_name': 'areacella',
'ensemble': 'r0i0p0',
'exp': 'historical',
'timerange': '1850/2000'}]
ESMValCore can automatically add extra facets based on the project, mip, short_name, and dataset. These extra facets are automatically added and used when searching for input files.
[5]:
tas.augment_facets()
tas
[5]:
Dataset:
{'dataset': 'MPI-ESM-MR',
'project': 'CMIP5',
'mip': 'Amon',
'short_name': 'tas',
'ensemble': 'r1i1p1',
'exp': 'historical',
'frequency': 'mon',
'institute': ['MPI-M'],
'long_name': 'Near-Surface Air Temperature',
'modeling_realm': ['atmos'],
'original_short_name': 'tas',
'product': ['output1', 'output2'],
'standard_name': 'air_temperature',
'timerange': '1850/2000',
'units': 'K'}
supplementaries:
{'dataset': 'MPI-ESM-MR',
'project': 'CMIP5',
'mip': 'fx',
'short_name': 'areacella',
'ensemble': 'r0i0p0',
'exp': 'historical',
'frequency': 'fx',
'institute': ['MPI-M'],
'long_name': 'Atmosphere Grid-Cell Area',
'modeling_realm': ['atmos', 'land'],
'original_short_name': 'areacella',
'product': ['output1', 'output2'],
'standard_name': 'cell_area',
'units': 'm2'}
session: 'session-686367c0-001c-4864-839d-c20887cf7415_20230301_160531'
Use the find_files method to find the files corresponding to the dataset.
[6]:
tas.find_files()
print(tas.files)
for supplementary_ds in tas.supplementaries:
print(supplementary_ds.files)
[ESGFFile:cmip5/output1/MPI-M/MPI-ESM-MR/historical/mon/atmos/Amon/r1i1p1/v20120503/tas_Amon_MPI-ESM-MR_historical_r1i1p1_185001-200512.nc on hosts ['aims3.llnl.gov', 'esgf.ceda.ac.uk', 'esgf.nci.org.au', 'esgf1.dkrz.de']]
[ESGFFile:cmip5/output1/MPI-M/MPI-ESM-MR/historical/fx/atmos/fx/r0i0p0/v20120503/areacella_fx_MPI-ESM-MR_historical_r0i0p0.nc on hosts ['aims3.llnl.gov', 'esgf.ceda.ac.uk', 'esgf.nci.org.au', 'esgf1.dkrz.de']]
If the files are not available locally, ESMValCore can download them for us.
[7]:
files = list(tas.files)
for supplementary_ds in tas.supplementaries:
files.extend(supplementary_ds.files)
files = [file for file in files if isinstance(file, ESGFFile)]
download(files, CFG['download_dir'])
tas.find_files()
print(tas.files)
for supplementary_ds in tas.supplementaries:
print(supplementary_ds.files)
[LocalFile('~/climate_data/cmip5/output1/MPI-M/MPI-ESM-MR/historical/mon/atmos/Amon/r1i1p1/v20120503/tas_Amon_MPI-ESM-MR_historical_r1i1p1_185001-200512.nc')]
[LocalFile('~/climate_data/cmip5/output1/MPI-M/MPI-ESM-MR/historical/fx/atmos/fx/r0i0p0/v20120503/areacella_fx_MPI-ESM-MR_historical_r0i0p0.nc')]
The data in the files can be loaded into an iris.cube.Cube. ESMValCore will automatically check for and fix problems with the data formatting and attach the cell area.
[8]:
cube = tas.load()
cube
[8]:
| Air Temperature (K) | time | latitude | longitude |
|---|---|---|---|
| Shape | 1812 | 96 | 192 |
| Dimension coordinates | |||
| time | x | - | - |
| latitude | - | x | - |
| longitude | - | - | x |
| Cell measures | |||
| cell_area | - | x | x |
| Scalar coordinates | |||
| height | 2.0 m | ||
| Cell methods | |||
| mean | time | ||
| Attributes | |||
| Conventions | 'CF-1.4' | ||
| associated_files | 'baseURL: http://cmip-pcmdi.llnl.gov/CMIP5/dataLocation gridspecFile: gridspec_atmos_fx_MPI-ESM-MR_historical_r0i0p0.nc ...' | ||
| branch_time | 0.0 | ||
| cmor_version | '2.6.0' | ||
| contact | 'cmip5-mpi-esm@dkrz.de' | ||
| experiment | 'historical' | ||
| experiment_id | 'historical' | ||
| forcing | 'GHG,Oz,SD,Sl,Vl,LU' | ||
| frequency | 'mon' | ||
| initialization_method | 1 | ||
| institute_id | 'MPI-M' | ||
| institution | 'Max Planck Institute for Meteorology' | ||
| model_id | 'MPI-ESM-MR' | ||
| modeling_realm | 'atmos' | ||
| parent_experiment | 'N/A' | ||
| parent_experiment_id | 'N/A' | ||
| parent_experiment_rip | 'N/A' | ||
| physics_version | 1 | ||
| product | 'output' | ||
| project_id | 'CMIP5' | ||
| realization | 1 | ||
| references | 'ECHAM6: n/a; JSBACH: Raddatz et al., 2007. Will the tropical land biosphere ...' | ||
| source | 'MPI-ESM-MR 2011; URL: http://svn.zmaw.de/svn/cosmos/branches/releases/mpi-esm-cmip5/src/mod; ...' | ||
| table_id | 'Table Amon (27 April 2011) a5a1c518f52ae340313ba0aada03f862' | ||
| title | 'MPI-ESM-MR model output prepared for CMIP5 historical' | ||
[9]:
cell_area = cube.cell_measures()[0]
cell_area
[9]:
<CellMeasure: cell_area / (m2) <lazy> shape(96, 192)>
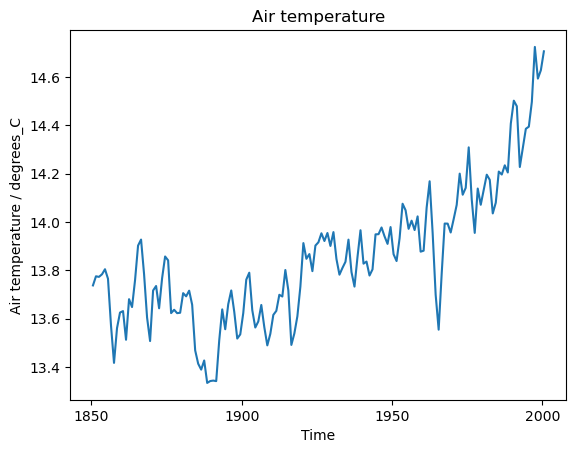
This code shows how to use some esmvalcore.preprocessor functions to compute the global annual mean temperature in degrees Celsius:
[10]:
cube = area_statistics(cube, operator='mean')
cube = annual_statistics(cube, operator='mean')
cube.convert_units('degrees_C')
The iris.quickplot module has useful functions for quickly plotting the results:
[11]:
iris.quickplot.plot(cube)
plt.show()