6. Running the ESMValTool¶
The following section gives a brief description of the steps required by a user to run an existing diagnostic. As an example, the toy diagnostic MyDiag is chosen to illustrate the basic steps:
Copy the template for the “namelist configuration file” (config_private_template.xml) to config_private.xml and adapt the path names within config_private.xml to your system structure (see Section 3.4). If needed, set/change the base path names for the input data (model and observations) and the output data. config_private.xml is ignored by version control (.gitignore) and should be backed up elsewhere for reuse. Note: the use of a “namelist configuration file” is optional in order to allow for machine specific standard search paths for input (and output) data without having to change the actual namelists. Alternatively, explicit path names can be used in the namelists.
Check/edit the main namelist nml/namelist_MyDiag.xml:
Set/check the file name of the “namelist configuration file” (defining the base path names) to be used by the ESMValTool (typically, this is the second line in the namelist, e.g., <include href=”./config_private.xml”/>).
If needed, set the pathnames in the <GLOBAL> section for the “work” directory (wrk_dir), the directory for the plots (plot_dir) and the directory for reformatted files (climo_dir). See Section 3.1 for details and Table 3.1 for a complete list of variables in the <GLOBAL> section.
In the <MODELS> section, define the model(s) to be used, including the root path for the actual model data, e.g.,
CMIP5_ETHZ MPI-ESM-LR Amon historical r1i1p1 2000 2004 @{MODELPATH}/ETHZ_CMIP5/
(see step 1 and Section 3.4 for details on how to set base path names such as @{MODELPATH}; alternatively, explicit path names can be used). See Section 3.2, Table 3.2 and Table 3.3 for details. The first year (here: 2000) and last year (here: 2004) of the model data processed for each model is specified in this section.
Optionally, change variable and the field type in the <DIAGNOSTICS> section. See Section 3.3, Table 3.5 and Table 3.6 for details. An overview of the available “field types” is given in Table 3.7, Table 3.8 lists the available variables. Please note that the diagnostic section may include additional models and/or observational data.
Check/edit the configuration file nml/cfg_MyDiag/cfg_MyDiag.ncl. In case of the toy diagnostics MyDiag, you can for example change the map projection for the contour plot by changing the value of the attribute diag_script_info@projection.
Run the ESMValTool (in the ESMValTool root directory): python main.py nml/namelist_MyDiag.nml
The output will be written to a subdirectory named like the diagnostics package (e.g., MyDiag) in the directories specified in the <GLOBAL> section of the namelist (see step 1 and also Section 3.4). The default directories are: work/MyDiag for the NetCDF output and work/plots/MyDiag for the plot(s) (see also Figure 6.1). Acknowledgements and references are written to the file work/refs-acknows_MyDiag.txt.
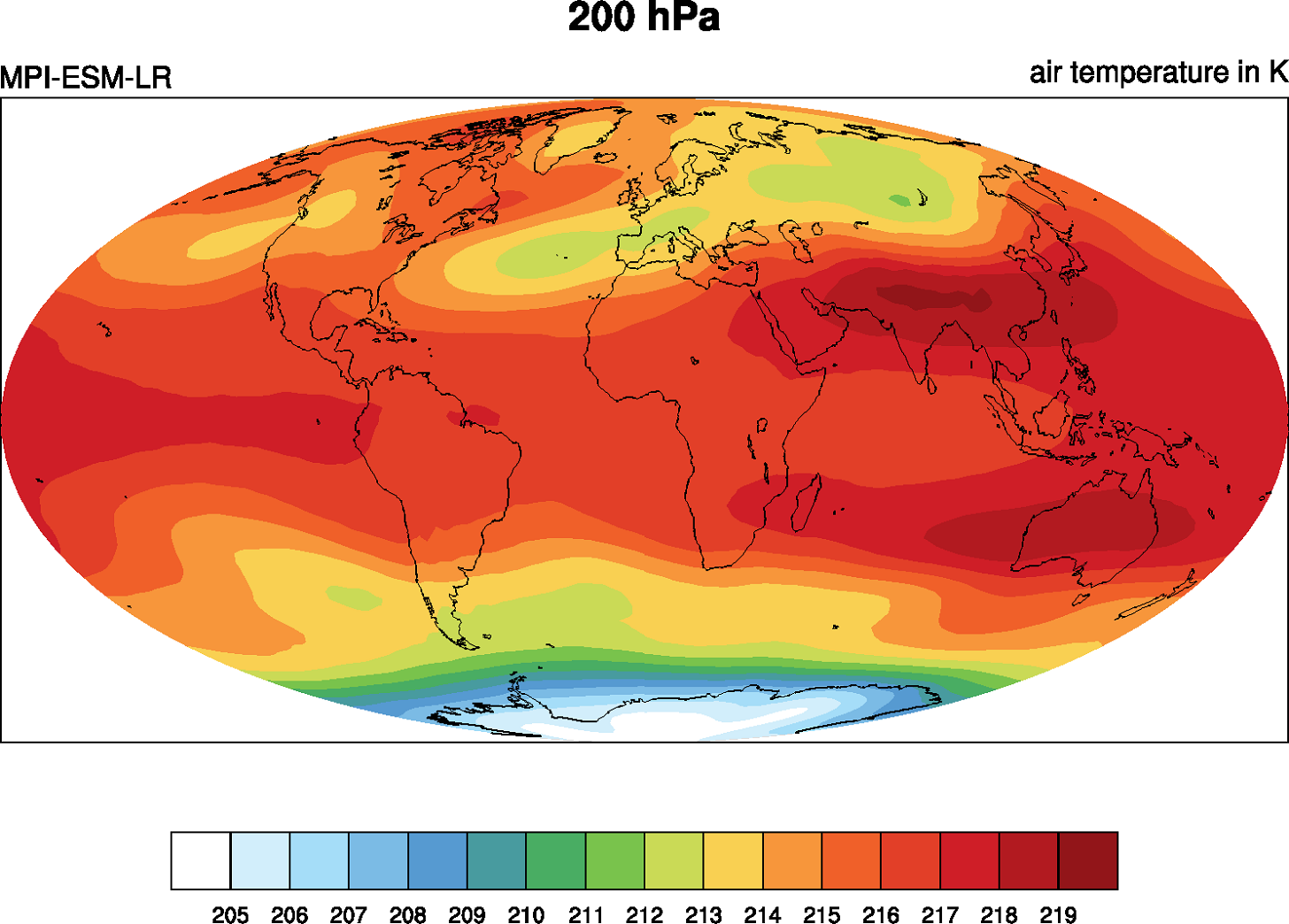
Figure 6.1 Example plot created by the toy diagnostic MyDiag showing the 5-year annual mean temperature at 200 hPa from the CMIP5 historical run (r1i1p1) with the MPI-ESM-LR model.
6.1. Available diagnostics and metrics¶
An introduction to the available diagnostics and metrics packages implemented into the ESMValTool v1.1 including a description of the user settings, observational data used, references, and example plots is given in Part annex_c.
6.2. Model and observational data¶
6.2.1. Model data¶
The project specifier (see Table 3.2) used in the <MODELS> section of the namelist (see Section 3.2 for details) determines the directory structure and file naming convention expected by the ESMValTool. The two most commonly used project specifiers are CMIP5 and CMIP5_ETHZ. Both are used to process CMIP5 data available from the Earth System Federation Grid (e.g., http://esgf.llnl.gov/). In order to download CMIP5 data, registration and creation of an “openID” is required. Instructions can be found here: http://cmip-pcmdi.llnl.gov/cmip5/data_getting_started.html.
Besides downloading files individually, the CMIP5 data portal is capable of generation a script for automated download of model data using GNU wget (https://www.gnu.org/software/wget/). The wget script generation is recommended for downloading a large number files and/or large data volume.
Any CMIP5 files downloaded to a single directory can be moved to a CMIP5 like directory structure using the NCL script util/CMIP5_sort/CMIP5_sort.ncl. The script CMIP5_sort.ncl expects all files to be moved to the CMIP5 like directory structure in the current directory (“.”). The root directory for creating the CMIP5 like directory structure is specified in the script CMIP5_sort.ncl via the variable “outpath”. The script is run via:
ncl <path of the ESMValTool>/util/CMIP5_sort/CMIP5_sort.ncl
The CMIP5 files will be moved into the directories outpath/experiment/mip/variable/name/ensemble/ for direct usage with the roject specifier CMIP5_ETHZ (see below). “experiment”, “mip”, “variable” and “name” (= model name) are automatically extracted from the filename.
The project specifier CMIP5
Syntax of the CMIP5 specifier in the <model>-tag (see Section 3.2 and Table 3.2 for details):
<model> CMIP5 name mip experiment ensemble start_year end_year path </model>
The project specifier CMIP5 will search for files in “path” with filenames matching the pattern
variable_mip_name_experiment_ensemble_.nc*
Note: “variable” is specified in the <diag>-section (see Section 3.3 for details). If “variable” is a derived quantity, all variables needed to calculate the derived quantity are processed automatically.
The project specifier CMIP5_ETHZ
Syntax of the CMIP5_ETHZ specifier in the <model>-tag (see Section 3.2 and Table 3.2 for details):
- <model> CMIP5_ETHZ name mip experiment ensemble start_year end_year
- path </model>
The project specifier CMIP5_ETHZ will search for files in “path/experiment/mip/variable/name/ensemble” with filenames matching the pattern
variable_mip_name_experiment_ensemble_*.nc
This directory structure resembles the ESGF CMIP5 directory structure.
Note: “variable” is specified in the <diag>-section (see Section 3.3 for details). If “variable” is a derived quantity, all variables needed to calculate the derived quantity are processed automatically.
6.2.2. Observational / reanalysis data¶
When possible, observations from the obs4MIPs/ana4MIPs archives are used in the model evaluation. These data are freely available from the ESGF in the same format as the CMIP simulations and can be directly used in the ESMValTool using the obs4mips or ana4mips project specifiers (see Table 3.2) in the namelist.
A collection of all observational data used by the diagnostics of the ESMValTool (MASTER BRANCH) is hosted at DLR and can be made available (restrictions by the data owner permitting) on request (see Table 6.1). The reformatted observational data can be read using e.g., the OBS class in the namelist (see below).
All observations are tiered as follows:
- Tier 1: data sets from the obs4MIPs and ana4MIPs archives
- Tier 2: other freely available data sets
- Tier 3: restricted data sets (e.g., license agreement required)
Observational data sets not available in the obs4MIPs/ana4MIPs archives need to be reformatted according to the CF/CMOR standard before they can be used (see Section 6.2.3 for more details).
The project specifier OBS
Syntax of the OBS specifier in the <model>-tag (see Section 3.2 and Table 3.2 for details):
- <model> OBS name case_name ensemble start_year end_year path
- </model>
The project specifier OBS will search for files in “path” with filenames matching the pattern
OBS_name_casename_ensemble_fieldtype_variable*.nc
Note: “variable” and “fieldtype” are specified in the <diag>-section (see Section 3.3 for details). If “variable” is a derived quantity, all variables needed to calculate the derived quantity are processed automatically.
The project specifier obs4mips
Syntax of the obs4mips specifier in the <model>-tag (see Section 3.2 and Table 3.2 for details):
- <model> obs4mips name process_level ensemble start_year end_year path
- </model>
The project specifier obs4mips will search for files in “path/name/” with filenames matching the pattern
variable_name_processlevel_ensemble_*.nc
Note: “variable” is specified in the <diag>-section (see Section 3.3 for details). If “variable” is a derived quantity, all variables needed to calculate the derived quantity are processed automatically.
6.2.3. Downloading and creating observational data sets¶
obs4MIPS and ana4MIPs data sets (“tier 1”, see above) are freely available from the ESGF. These data sets can be used directly with the ESMValTool without the need for reformatting. Examples of such data sets include:
- AIRS
- CERES-EBAF
- CFSR
- CloudSat-L3
- GPCP-1DD, GPCP-SG
- IFS-Cy31r2
- ISCCP
- MERRA
- MISR
- MODIS
- TES
- TRMM, TRMM-L3
For the required file naming conventions and the expected directory structure see Section :numref:`obs_data`
For all other (non-obs4MIPs and non-ana4MIPs) data sets, reformatting routines are provided with downloading and processing instructions in the header of the scripts. These reformatting scripts can be found in:
reformat_scripts/obs/
These reformat scripts can be specified in a namelist-file (e.g. namelist_reformat_obs.xml) and executed by calling the main.py script with the option “-r”:
python main.py -r namelist_reformat_obs.xml
This reformat namelist file contains the tag <REFORMAT> that can hold multiple <reformat_script>-tags specifying the reformat scripts to be called:
<REFORMAT>
<reformat_script> /PATH/TO/REFORMATSCRIPT</reformat_script>
</REFORMAT>
An example reformat namelist file is listed below:
<namelist>
<include href="./config_private.xml"/>
<namelist_summary>
###############################################################################
namelist_reformat_obs.xml
Description
Special namelist for reformatting observational data.
The currently available reformat scripts are stored in reformat_scripts/obs/
To run this namelist the -r option must be given:
python main.py -r nml/namelist_reformat_obs.xml
This namelist is part of the ESMValTool
###############################################################################
</namelist_summary>
<REFORMAT>
<reformat_script id=obs_1>./reformat_scripts/obs/reformat_obs_1.ncl </reformat_script>
<reformat_script id=obs_2>./reformat_scripts/obs/reformat_obs_2.ncl </reformat_script>
<reformat_script id=obs_3>./reformat_scripts/obs/reformat_obs_3.ncl </reformat_script>
<reformat_script id=obs_N>./reformat_scripts/obs/reformat_obs_N.ncl </reformat_script>
</REFORMAT>
</namelist>
A list of available data sets and their corresponding reformatting routines are given in Table 6.1.
Table 6.1 Observational data for use with the ESMValTool. See headers of the reformatting routines for downloading and processing instructions.
| Name | Tier | Description | Variables | Type | Time range | Script name |
|---|---|---|---|---|---|---|
| ACCESS | 3 | Aerosol vertical profiles | mmrbc | Campaign | – | reformat_obs_ACCESS.ncl |
| ACCESS-2 | 3 | Aerosol vertical profiles | conccnd5, conccnd10 | Campaign | 2014-2014 | reformat_obs_ACCESS-2.ncl |
| AERONET | 2 | Aerosol optical depth at 550nm | od550aer | Ground | 1992-2012 | reformat_obs_AERONET.ncl |
| AIRS | 1 | relative humidity, temperature | hur, hus, ta | Satellite | 2003-2010 | none (obs4MIPS) |
| Asmi11 | 2 | Aerosol size distributions | sizecnSTP | Ground | 2009-2010 | reformat_obs_Asmi11.ncl |
| AURA-MLS-OMI | 2 | Tropospheric column ozone | tropoz | Satellite | 2005-2013 | reformat_obs_AURA-MLS-OMI.ncl |
| AURA-TES | 2 | Ozone mixing ration | vmro3 | Satellite | 2005-2009 | reformat_obs_AURA-TES.ncl |
| BDBP | 3 | zonally averaged ozone profiles | tro3prof | Ozone sondes | 1979-2007 | reformat_obs_BDBP.ncl |
| CARSNET | 2 | Aerosol optical depth at 550 nm | od550aer | Ground | 2002-2013 | reformat_obs_CARSNET.ncl |
| CASTNET | 2 | Aerosol surface level concentration | concso4, concso3, concnh4 | Ground | S1987-2012 | reformat_obs_CASTNET.ncl |
| CERES | 3 | CERES synoptic data (radiative fluxes at surface, toa) | rsuscs, rsus, rsdscs, rsds, rluscs, rlus, rldscs, rlds, rsutcs, rsut, rlutcs, rlut | Satellite | 2004 | reformat_obs_CERES-SYN1deg -SFC.bash, reformat_obs_CERES- SYN1deg-TOA.bash |
| CFSR | 1 | Surface pressure | psl | Reanalysis | 2013 | none (obs4MIPs) |
| CIRRUS | 3 | Aerosol vertical profiles | mmrbc, mmrbcfree | Campaign | late Nov. 2006 | reformat_obs_CIRRUS.ncl |
| CLARA-A2 | 2 | Cloud cover | clt | Satellite | 1982-2015 | contact ESMValtool development team |
| CloudSat | 1 | Cloud cover | clt | Satellite | 2006-2010 | reformat_obs_cloudsat.bash |
| CMAP | 2 | Precipitation | pr | merged analysis | 1980-2013 | reformat_obs_CMAP.ncl |
| Concert | 3 | Aerosol vertical profiles | mmrbc, conccnSTP14 | Campaign | – | reformat_obs_CONCERT.ncl |
| CR-AVE | 3 | Aerosol vertical profiles | mmrbc | Campaign | – | reformat_obs_CR-AVE.ncl |
| CRU | 3 | Surface temperature, precipitation | tas, pr | Reanalysis | 1901-2006 | reformat_obs_CRU.ncl |
| DC3 | 3 | Aerosol vertical profiles | mmrbc | Campaign | – | reformat_obs_DC3.ncl |
| Dong08-ARGO | 2 | Derived ocean mixed layer depth | mlotst | Campaign | 2001-2006 | reformat_obs_Dong08-ARGO- monthly.ncl |
| EANET | 3 | Aerosol surface level concentrations | concso4, consco3, concnh4 | Ground | 2001-2005 | reformat_obs_EANET.ncl |
| EMEP | 2 | Aerosol surface level concentration | concso4, concno3, concnh4, concnh4, concpm2p5, concpm10 | Ground | 1970-2012 | reformat_obs_EMEP.csh |
| Emmons | 2 | Vertical profiles of gases | various | Campaign | variable | reformat_obs_Emmons.csh |
| ERA-40 | 3 | essential climate variables | ta, ua | Reanalysis | 1960-2001 | |
| ERA-Interim | 3 | Basic climate parameters | ta, ua, va, zg, hus, tas, tos, ps, psl, tauu, tauv, clwvi, clivi, sftif | Reanalysis | 1979-2012 | reformat_obs_ERA-Interim.ncl, reformat_obs_ERA-Interim- surffluxes.ncl |
| ERA-Interim fluxes | 3 | Basic climate parameters, surface fluxes | pr, evspsbl, hfls, hfss, rsns, rlns | Forecast | 2000-2005 | reformat_obs_ERA-Interim-surffl uxes.ncl |
| ESACCI-AEROSOL | 2 | Aerosol optical depth at 550 nm | od550aer, od870aer, od550lt1aer, abs550aer, od550aer-Stderr, od870aer-Stderr | Satellite | 1997-2011 | reformat_obs_ESACCI-AEROSOL.ncl |
| ESACCI-CLOUD | 2 | Total cloud fraction, Liquid water path, Ice water path | clt, clwvi, clivi, lwpStderr, iwpStderr , cltStderr | Satellite | 2007-2009 | reformat_obs_ESACCI-CLOUD.ncl |
| ESACCI-GHG | 2 | column averaged CO2and CH4 | xco2, xco2Stderr, xch4, xch4Stderr | Satellite | 2003-2014 | reformat_obs_ESACCI-GHG.ncl |
| ESACCI-OZONE | 2 | Total ozone column, Tropospheric column ozone, Ozone mixing ratio | toz, tro3prof, tozStderr, tro3Stderr | Satellite | 2007-2008 | reformat_obs_ESACCI-OZONE.ncl, reformat_obs_ESACCI-OZONE- PL.ncl |
| ESACCI-SIC | 2 | Sea ice concentrationtoz | sic, sicStderr | Satellite | 2003-2010 | reformat_obs_ESACCI-SIC.ncl |
| ESACCI- SOILMOISTURE | 2 | Degree of saturation | dos, dosStderr, sm, smStderr | Satellite | 1988-2008 | reformat_obs_ESACCI- SOILMOISTURE.ncl |
| ESACCI-SST | 2 | Sea surface temperature (saved as skin temperature) | ts, tsStderr | Satellite/ Analysis | 1992-2010 | reformat_obs_ESACCI-SST.ncl |
| ESRL | 2 | CO2 surface level concentrations | co2 | Ground | 1973-2012 | reformat_obs_ESRL.ncl |
| ETH-SOM-FFN | 2 | pCO2 ocean surface | spco2 | – | 1998-2011 | reformat_obs_ETH-SOM-FFN.ncl |
| GCP | 2 | CO2 exchange | co2flux, fgco2, nbp | Reanalysis | 1959-2011 | reformat_obs_GCP.ncl |
| GLOBAL-VIEW | 2 | CO surface level concentrations | vmrco | Ground | 1991-2008 | reformat_obs_GLOBAL-VIEW.ncl |
| GPCC | 2 | Precipitation | pr | Reanalysis | 1901-2010 | reformat_obs_GPCC.ncl |
| GPCP | 1 | Precipitation | pr, prStderr | – | 1979-2013 | none (obs4MIPs) |
| GTO-ECV | 3 | Total column ozone | toz | Satellite | 1996-2010 | reformat_obs_GTO-ECV.ncl |
| HadCRUT | 2 | Near-surface air temperature | tas | Ground | 1850-2013 | reformat_obs_HadCRUT.ncl reformat_obs_HadCRUT4.ncl |
| HadISST | 2 | Sea ice concentrations and sea surface temperatures | sic, ts | Reanalysis | 1870-2014 | reformat_obs_HadISST.ncl |
| HALOE | 2 | Water vapour mixing ratio | vmrh2o | Satellite | 1991-2002 | reformat_obs_HALOE.ncl |
| HIPPO | 3 | Aerosol vertical profiles | mmrbc | Campaign | – | reformat_obs_HIPPO.ncl |
| HWSD | 2 | Soil carbon content | cSoil | Ground | 2000 | |
| IFS-Cy31r2 | 1 | Surface pressure | psl | Reanalysis | 1979-2013 | none (obs4MPIs) |
| IMPROVE | 2 | Aerosol surface level concentrations | concso4, concno3, concnh4, concbc, concoa, concpm2p5, concpm10 | Ground | 1988-2011 | reformat_obs_IMPROVE.ncl |
| INCA | 3 | Aerosol vertical profiles | conccnSTP5, conccnSTP14, conccnSTP120 | Campaign | – | reformat_obs_INCA.ncl |
| ISCCP | 1 | Cloud properties | albisccp, clisccp, cltisccp, cttisccp | Satellite | 1984-2007 | none (obs4MPIs) |
| ISCCP-FD-SRF | 2 | Clear-sky radiative fluxes | rsdscs, rsuscs | Satellite | 1984-2009 | |
| JMA-TRANSCOM | 3 | CO2 exchange | nbp, fgco2 | Reanalysis | 1985-2008 | |
| LACE | 2 | Aerosol size distributions | sizecn | Campaign | – | reformat_obs_LACE.ncl |
| LAI3g | 3 | Leaf area index | LAI | Reanalysis | 1982-2010 | |
| LandFlux-EVAL | 3 | Evapotranspi-ration | et, et-sd | Synthesis product (model + observa- tions) | 1989-2005 | reformat_obs_landflux-eval.ncl |
| MERRA | 1 | Precipitation | pr | Reanalysis | 1979-2011 | none (obs4MPIs) |
| MISR | 1 | Aerosol optical depth | od550aer | Satellite | 2001-2012 | none (obs4MPIs) |
| MLS | 1 | humidity, temperature | hus, husStderr, ta, taStderr | Satellite | 2005-2010 | none (obs4MPIs) |
| MODIS-CFMIP | 2 | Ice water path | clivi | Satellite | 2003-2014 | reformat_obs_MODIS-CFMIP.ncl |
| MODIS_ L3_C6 | 2 | Ice water path, liquid water path, total cloud cover, aerosol optical depth | clivi, clwvi, clt, od550aer | Satellite | 2003-2014 | reformat_obs_MODIS-L3-C6.ncl |
| MTE | 2 | Gross primary productivity of carbon | gpp | Reanalysis | 1982-2008 | |
| NCEP | 2 | Essential climate variables | ta, ua, va, zg, hus, tas | Reanalysis | 1948-2012 | reformat_obs_NCEP.ncl, reformat_obs_NCEP-daily.ncl |
| NDP | 2 | Vegetation carbon content | cVeg | Ground | 2000 | |
| NIWA | 3 | Total column ozone | toz | Reanalysis | 1980-2010 | reformat_obs_NIWA.ncl |
| NOAA interpola- ted OLD | 2 | Interpolated outgoing longwave radiation | rlut | Satellite | 1975-2013 | reformat_obs_NOAA-PSD- Interp.ncl |
| NSIDC | 2 | Sea ice concentrations | sic | Satellite | 1978-2010 | reformat_obs_NSIDC.ncl |
| PATMOS | 2 | Cloud cover | clt | Satellite | 1982-2014 | contact ESMValtool development team |
| Putaud | 2 | Aerosol size distributions | sizecn | Campaign | – | reformat_obs_Putaud.ncl |
| SALTRACE | 3 | Aerosol vertical profiles | mmrbc | Campaign | – | reformat_obs_SALT-RACE.ncl |
| SeaWIFS | 2 | Ocean biochemistry | chl | 1997-2010 | reformat_obs_SeaWIFS.csh | |
| SOCAT | 2 | Ocean surface CO2 | spco2 | 1970-2011 | reformat_obs_SOCAT.csh | |
| SRB | 2 | Radiative fluxes | rsut, rlut, rlutcs | Satellite | 1983-2007 | reformat_obs_SRB.ncl |
| SSMI-MERIS | 1 | Water vapour path | prw, prwStderr | Satellite | 2003-2008 | none (obs4MIPs) |
| takahashi14 | 2 | Ocean biogeochemistry | talk | 2005 | reformat_obs_takahashi14.csh | |
| TC4 | 3 | Aerosol vertical profiles | mmrbc | Campaign | – | reformat_obs_TC4.ncl |
| TES | 1 | Ozone | tro3 | 2006-2009 | reformat_obs_TES.ncl | |
| Texas | 3 | Aerosol vertical profiles | mmrbc, mmraer | Campaign | – | reformat_obs_Texas.ncl |
| Tilmes | 2 | Ozone mixing ratios | vmro3 | in-situ | 1995-2009 | reformat_obs_Tilmes.ncl |
| TOMS | 2 | Total ozone column | toz | Satellite | 1990 | |
| TRMM-3B42 | 2 | Precipitation | pr | Satellite | 1998-2014 | reformat_obs_TRMM-3B42- daily.ncl, reformat_obs_TRMM- 3B42-3hourly.ncl |
| UCN-Pacific | 3 | Aerosol vertical profiles | conccnSTP3 | Campaign | – | reformat_obs_UCN-Pacific.ncl |
| UWisc | 3 | Liquid water path | clwvi, lwpStderr | Satellite | 1988-2007 | reformat_obs_UWisc.ncl |
| WHOI-OAFlux | 2 | Global ocean heat flux and evaporation | hfls, hfss | Analysis | 1958-2013 | reformat_obs_WHOI-OAFlux.ncl |
| WOA09 | 2 | Climatological ocean fields | so, sos, to, tos | Analyzed climatology | – | reformat_obs_WOA09.ncl |
| woa2005 | 2 | Ocean biogeochemistry | o2 | 2005 | reformat_obs_woa2005.csh |
6.3. The acknowledgements log file¶
Each diagnostics in the tool automatically generates a log file containing a list of authors/contributors, details on the projects to be acknowledged and the reference papers to be cited. It also provides a list of the used model and observational data with the corresponding references.
The log is created automatically when running the ESMValTool. The log file is named refs-acknow_<diagnostics>.txt and written to the directory defined in the <GLOBAL> section of the namelist (variable wrk_dir, see Section 3.1), e.g., work/refs-acknows_MyDiag.txt (see also Section 6, step 4).
An example excerpt of an acknowledgements log file is provided below.
Example
---------------------------------------------------------------------------
+++++++++++++ ESMValTool REFERENCES and ACKNOWLEDGEMENTS LOG ++++++++++++++
---------------------------------------------------------------------------
Namelist file: namelist_perfmetrics_CMIP5.xml
Creation date: Wed Dec 16 22:58:29 CET 2016
ESMValTool version: 1.1.0
Host name: ###
User name: ###
Licensed under the Apache License, Version 2.0 (the "License"); you may
not use this file except in compliance with the License. You may obtain
a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS"BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
Please acknowledge the use of the ESMValTool.
Please cite Eyring et al., ESMValTool (v1.0) -- a community diagnostic and
performance metrics tool for routine evaluation of Earth System Models in
CMIP, Geosci. Model Dev., 2016.
For the specific diagnostics, see below.
===========================================================================
=== perfmetrics_main.ncl ===
AUTHOR(S):
-A- ###
CONTRIBUTOR(S):
-C- ###
-C- ###
-C- ###
REFERENCE(S) FOR THIS DIAGNOSTIC:
-R- Please cite Righi et al., Geosci. Model Dev., 8, 733-768
doi:10.5194/gmd-8-733-2015, 2015.
-R- Please cite Gleckler et al., J. Geophys. Res., 113, D06104,
doi:10.1029/2007JD008972, 2008.
REFERENCE(S) FOR THE OBSERVATIONS:
-R- NCEP - Kalnay et al., Bull. Amer. Meteor. Soc., 77, 437-470, 1996.
-R- ERA-Interim
-R- AIRS
-R- CERES-EBAF
-R- SRB
ACKNOWLEDGEMENTS FOR THE PROJECTS:
-P- EU FP7 project EMBRACE
-P- DLR project ESMVal
PREPROCESSING/REFORMATTING (ESMValTool v1.1.0):
Variable: ta
Model: ERA-Interim
Input file(s):
(1) OBS_reanaly_ERA-Interim_1_T3M_ta_2000-2001.nc
Original source file(s) of all input file(s):
-S- (1)
\@{OBSPATH}/Tier3/ERA-Interim/OBS_ERA-Interim_reanaly_1_T3M_ta.nc
Fixes applied to original source file(s): none
Reference(s) of original source file(s):
(1) Dee, D. P. et al., Q. J. Roy. Meteor. Soc., 137, 553-597,
doi:10.1002/qj.828, 2011.
Model: MPI-ESM-LR
Input file(s):
(1) CMIP5_ETHZ_Amon_historical_MPI-ESM-LR_r1i1p1_T3M_ta_1998-2002.nc
Original source file(s) of all input file(s):
-S- (1) \@{MODELPATH}/ETHZ_CMIP5/historical/Amon/ta/MPI-ESM-LR/r1i1p1/
ta_Amon_MPI-ESM-LR_historical_r1i1p1_199001-199912.nc
(tracking_id: ea695cd3-6234-4ddf-a68e-b4d82a2e7305)
-S- (2) \@{MODELPATH}/ETHZ_CMIP5/historical/Amon/ta/MPI-ESM-LR/r1i1p1/
ta_Amon_MPI-ESM-LR_historical_r1i1p1_200001-200512.nc
(tracking_id: f9134520-0445-4461-9a48-14d8663dab74)
Fixes applied to original source file(s):
./reformat_scripts/fixes/CMIP5_MPI-ESM-LR_fix.ncl
[...]